
|
HOME
RESEARCH
LAB MEMBERS
PUBLICATIONS
CONTACT US
LINKS
|
|
|
Research in our lab focuses on two major areas: glia and their roles in the nervous system, and the control of programmed cell death during animal development.
In addition, we have developed tools for temporal control of gene expression in C. elegans, described strategies for optimizing genetic screens in the animal, and developed software to analyze polymorphisms in the C. elegans genome to facilitate gene cloning. Descriptions of each area are provided below.
Glial roles in the development and function of the nervous system 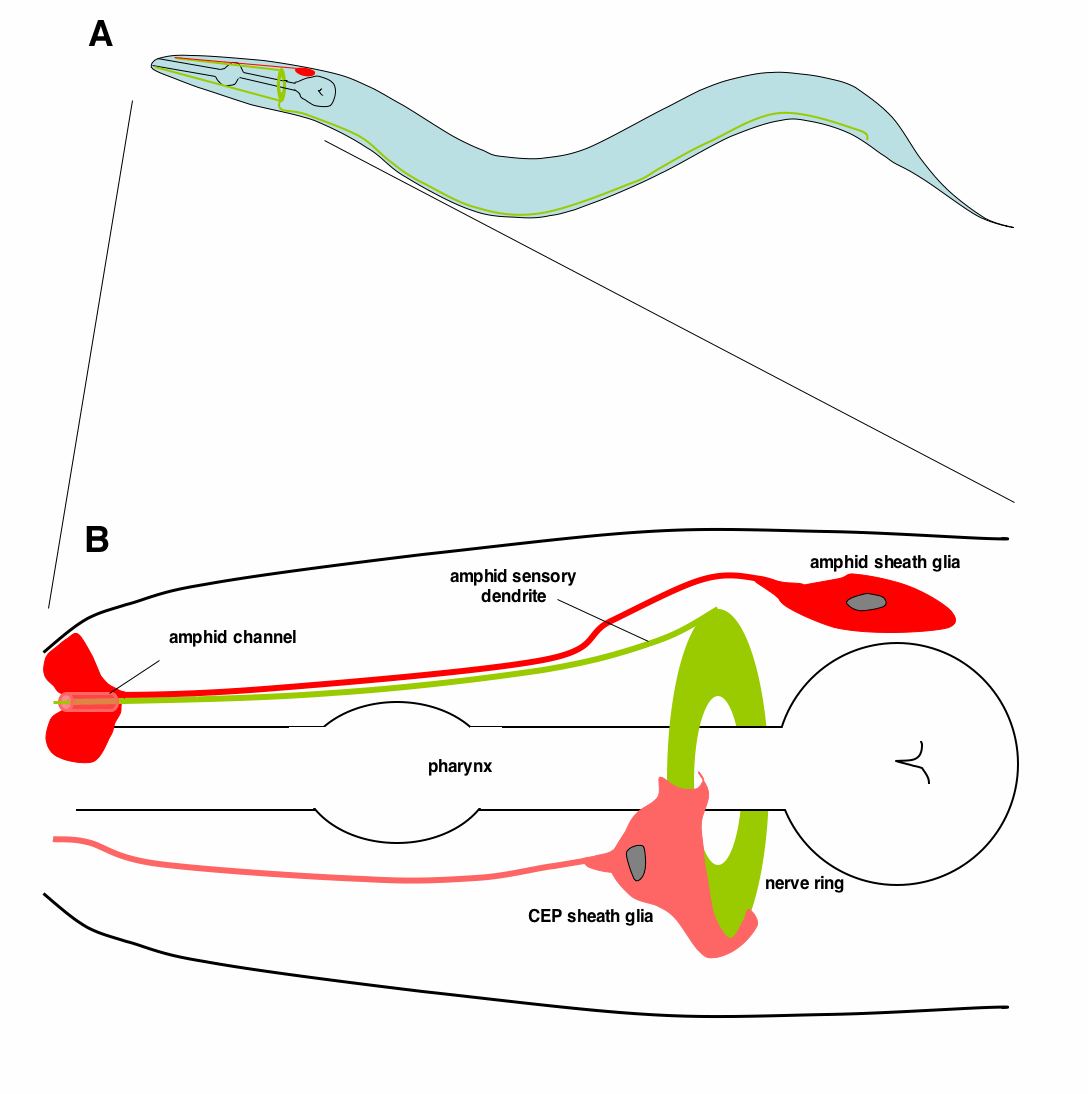 current propagation, and are likely to play key roles in many, if
not all aspects of nervous system function and development. C. elegans possesses 302 neurons and 56
glial cells. These glia fall into three broad categories: 50 glia associate with dendrites of sensory
neurons; of these, four (CEP sheath glia) also envelop the C. elegans nerve ring (its brain) and send
processes to some synaptic sites in the brain (see Figure). Six glia (GLR glia) form gap junctions to both
muscle cells and motor neurons, and are thus well-situated to regulate neuromuscular activity. We believe that C. elegans offers a unique opportunity to understand glial biology.
Specifically, in vertebrates and in Drosophila, in vivo (and often in vitro) removal of glia promotes the
death of associated neurons, preventing more subtle yet crucial aspects of glial influence on neuronal
activity to be studied, explaining to some extent why research on these cells has been comparatively limited.
Using cell ablation strategies we showed that in C. elegans, all glia we examined are not required for
survival (Yoshimura et al., 2008; Bacaj et al., 2008). This simple, yet powerful observation, allowed us to uncover important glial
functions that have not been previously demonstrated in vivo or at all.
current propagation, and are likely to play key roles in many, if
not all aspects of nervous system function and development. C. elegans possesses 302 neurons and 56
glial cells. These glia fall into three broad categories: 50 glia associate with dendrites of sensory
neurons; of these, four (CEP sheath glia) also envelop the C. elegans nerve ring (its brain) and send
processes to some synaptic sites in the brain (see Figure). Six glia (GLR glia) form gap junctions to both
muscle cells and motor neurons, and are thus well-situated to regulate neuromuscular activity. We believe that C. elegans offers a unique opportunity to understand glial biology.
Specifically, in vertebrates and in Drosophila, in vivo (and often in vitro) removal of glia promotes the
death of associated neurons, preventing more subtle yet crucial aspects of glial influence on neuronal
activity to be studied, explaining to some extent why research on these cells has been comparatively limited.
Using cell ablation strategies we showed that in C. elegans, all glia we examined are not required for
survival (Yoshimura et al., 2008; Bacaj et al., 2008). This simple, yet powerful observation, allowed us to uncover important glial
functions that have not been previously demonstrated in vivo or at all.
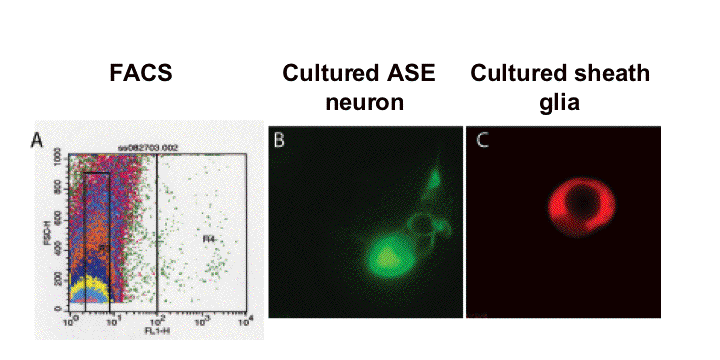 expression of specific gene products exclusively within glia. Glia-specific promoters were obtained in three ways. First, we surveyed the literature for genes whose expression patterns suggested they might be expressed in glia. Second, we cloned the daf-6 gene and showed that its promoter, and that of proteins similar to it in sequence, could drive expression in glia (Perens and Shaham, 2005; Yoshimura et al., 2008). Third, we dissociated embryos expressing GFP in the amphid sheath glial cell, FACS sorted GFP positive cells, and prepared mRNA, which was used to probe a C. elegans gene microarray. From this study we identified over 300 glia-enriched transcripts, of which we have confirmed over 20 by GFP fusion studies (Bacaj et al., 2008). This study has not only been useful for generating glial reagents, but has been instrumental in defining a set of candidate genes that may mediate glial functions.
expression of specific gene products exclusively within glia. Glia-specific promoters were obtained in three ways. First, we surveyed the literature for genes whose expression patterns suggested they might be expressed in glia. Second, we cloned the daf-6 gene and showed that its promoter, and that of proteins similar to it in sequence, could drive expression in glia (Perens and Shaham, 2005; Yoshimura et al., 2008). Third, we dissociated embryos expressing GFP in the amphid sheath glial cell, FACS sorted GFP positive cells, and prepared mRNA, which was used to probe a C. elegans gene microarray. From this study we identified over 300 glia-enriched transcripts, of which we have confirmed over 20 by GFP fusion studies (Bacaj et al., 2008). This study has not only been useful for generating glial reagents, but has been instrumental in defining a set of candidate genes that may mediate glial functions.
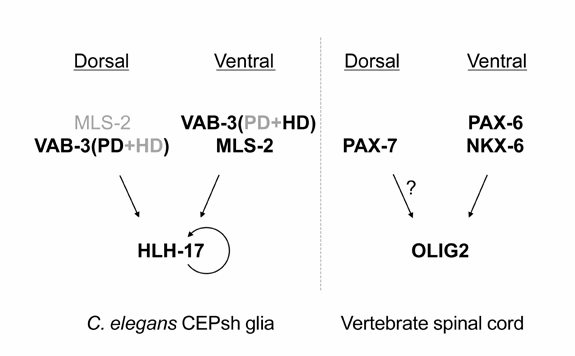 connected by slender processes to glial cell bodies. Our analysis of genes expressed in C. elegans glia has revealed that the 56 cells we have defined as glia define a cell class by their pattern of gene expression. We identified genes expressed nearly exclusively in all three glial types, or in subsets of these glial types. These observations support the anatomical/morphological classification of these cells as a group. In vertebrates, the Olig1/2 bHLH transcriptional regulators are essential for oligodendrocyte formation, and are also expressed in other glial types, suggesting that they are key regulators of glial cell fate. We compared the Olig1/2 sequences to the C. elegans protein database and identified a protein, HLH-17, that was most similar to Olig1/2. Remarkably, we found that hlh-17 was expressed strongly in all C. elegans glia, except the GLR glia (Yoshimura et al., 2008). From a genetic screen aimed at identifying regulators of hlh-17 expression in the CEP sheath glia we isolated mutants in a Pax7-like gene and a Nkx-like transcriptional regulator, which affect dorsal and ventral hlh-17 expression, respectively (Yoshimura et al., 2008). Remarkably, the Nkx6.1 protein in vertebrates controls Olig2 expression in the ventral spinal cord, and Pax7 may regulate Olig2 expression in dorsal spinal cord progenitor cells (see Figure).
Taken together, the results described here suggest a deep similarity between C. elegans and vertebrate glia, and suggest that understanding the functions of these cells in C. elegans may have direct relevance to understanding glia in all animals.
connected by slender processes to glial cell bodies. Our analysis of genes expressed in C. elegans glia has revealed that the 56 cells we have defined as glia define a cell class by their pattern of gene expression. We identified genes expressed nearly exclusively in all three glial types, or in subsets of these glial types. These observations support the anatomical/morphological classification of these cells as a group. In vertebrates, the Olig1/2 bHLH transcriptional regulators are essential for oligodendrocyte formation, and are also expressed in other glial types, suggesting that they are key regulators of glial cell fate. We compared the Olig1/2 sequences to the C. elegans protein database and identified a protein, HLH-17, that was most similar to Olig1/2. Remarkably, we found that hlh-17 was expressed strongly in all C. elegans glia, except the GLR glia (Yoshimura et al., 2008). From a genetic screen aimed at identifying regulators of hlh-17 expression in the CEP sheath glia we isolated mutants in a Pax7-like gene and a Nkx-like transcriptional regulator, which affect dorsal and ventral hlh-17 expression, respectively (Yoshimura et al., 2008). Remarkably, the Nkx6.1 protein in vertebrates controls Olig2 expression in the ventral spinal cord, and Pax7 may regulate Olig2 expression in dorsal spinal cord progenitor cells (see Figure).
Taken together, the results described here suggest a deep similarity between C. elegans and vertebrate glia, and suggest that understanding the functions of these cells in C. elegans may have direct relevance to understanding glia in all animals.
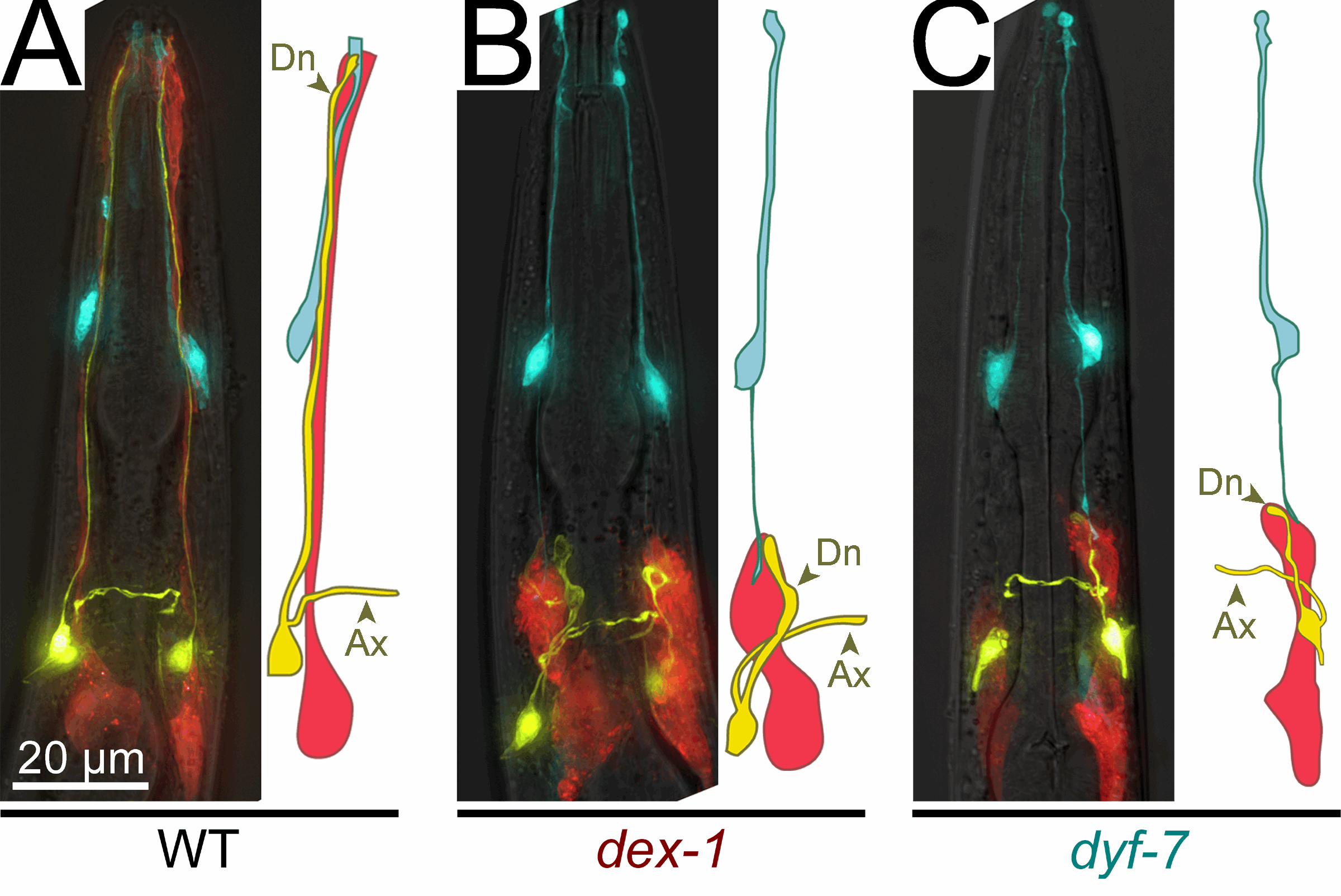 The amphid sensory organ is composed of two glial cells termed sheath and socket cells that associate with the dendritic endings of 12 sensory neurons. Ablation of the amphid sheath glia precursor resulted in dramatically shortened amphid neuron dendrites, suggesting that glia are important for dendrite extension (T. Bacaj, unpublished results). To study this phenotype in greater depth we performed a mutant screen to search for animals in which dendrite extension is defective (Heiman and Shaham, 2009). We identified 15 mutants, defining at least three different genes. We cloned two of these genes, dyf-7 and dex-1 (see Figure). DYF-7 protein is a transmembrane Zona Pellucida (ZP) domain protein similar to mammalian beta tectorin, a protein forming the tectorial membrane in the vertebrate cochlea, and to mammalian oocyte Zona Pellucida proteins. DYF-7 is expressed in amphid neurons, functions during dendrite extension in the embryo, and localizes to dendritic tips. dex-1 and dyf-7 exhibit cold-sensitive synergistic
The amphid sensory organ is composed of two glial cells termed sheath and socket cells that associate with the dendritic endings of 12 sensory neurons. Ablation of the amphid sheath glia precursor resulted in dramatically shortened amphid neuron dendrites, suggesting that glia are important for dendrite extension (T. Bacaj, unpublished results). To study this phenotype in greater depth we performed a mutant screen to search for animals in which dendrite extension is defective (Heiman and Shaham, 2009). We identified 15 mutants, defining at least three different genes. We cloned two of these genes, dyf-7 and dex-1 (see Figure). DYF-7 protein is a transmembrane Zona Pellucida (ZP) domain protein similar to mammalian beta tectorin, a protein forming the tectorial membrane in the vertebrate cochlea, and to mammalian oocyte Zona Pellucida proteins. DYF-7 is expressed in amphid neurons, functions during dendrite extension in the embryo, and localizes to dendritic tips. dex-1 and dyf-7 exhibit cold-sensitive synergistic
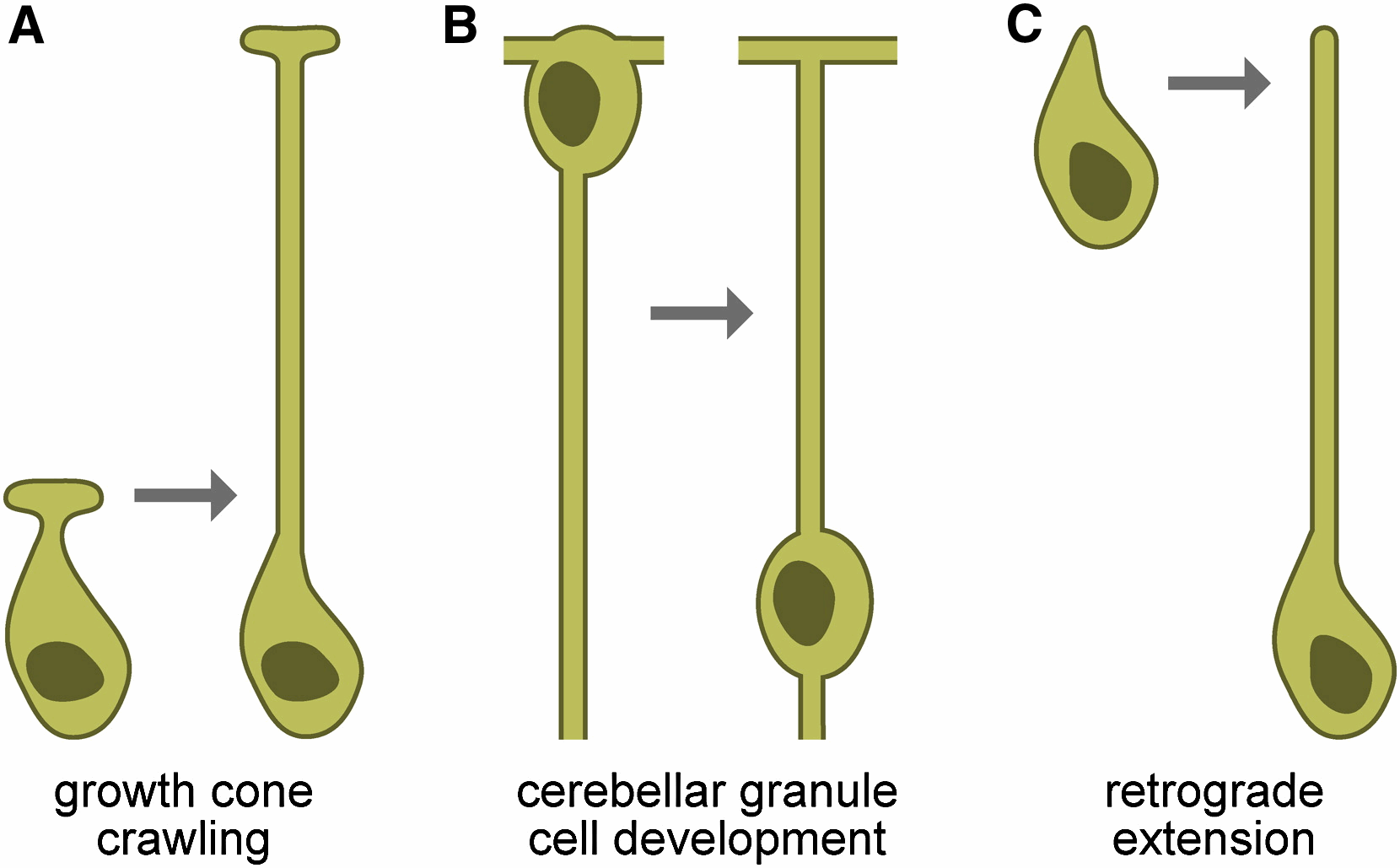 genetic interactions, suggesting that the two proteins might interact. Remarkably, DEX-1 is similar to alpha tectorin, a tectorial membrane protein also similar to sperm Zonadhesin protein. DEX-1 is also required during dendrite extension and is expressed in non-neuronal cells, and perhaps glia. Remarkably, using live imaging in embryos using photoconvertible reporters, we demonstrated that dendrites do not emanate from the cell body and extend. Rather, a dendritic bud anchors using DEX-1 and DYF-7, and the dendrite is extended as the cell body then migrates away. We call this novel mode of dendrite formation "retrograde extension" (see Figure).
genetic interactions, suggesting that the two proteins might interact. Remarkably, DEX-1 is similar to alpha tectorin, a tectorial membrane protein also similar to sperm Zonadhesin protein. DEX-1 is also required during dendrite extension and is expressed in non-neuronal cells, and perhaps glia. Remarkably, using live imaging in embryos using photoconvertible reporters, we demonstrated that dendrites do not emanate from the cell body and extend. Rather, a dendritic bud anchors using DEX-1 and DYF-7, and the dendrite is extended as the cell body then migrates away. We call this novel mode of dendrite formation "retrograde extension" (see Figure).We have also examined glial ensheathment of neurons during development by studying the daf-6 gene (Perens and Shaham, 2005). We showed that DAF-6 defines a new class of Patched-related proteins conserved in Drosophila and vertebrates. DAF-6 localizes to glial membranes that surround a subset of amphid dendrites and mutations in daf-6 display defects in forming the glial channel through which sensory neurons project. Animals in which dendritic sensory endings are defective (such as dyf-13 mutants we characterized; Blacque et al., 2005) mislocalize glial DAF-6 and have abnormal glial ensheathment, suggesting that a neuronal signal impinges on DAF-6 to allow proper formation of the glial sheath. DAF-6 is expressed in and affects most tubular structures in C. elegans, uncovering a hitherto unsuspected possible connection between neuronal ensheathment and tubulogenesis. Finally, we have identified a kinase module, containing the LIT-1 kinase that opposes the functions of DAF-6, and restricts tubulogenesis, suggesting that the interplay between DAF-6 and LIT-1 is crucial for establishing luminal diameter of the amphid channel. 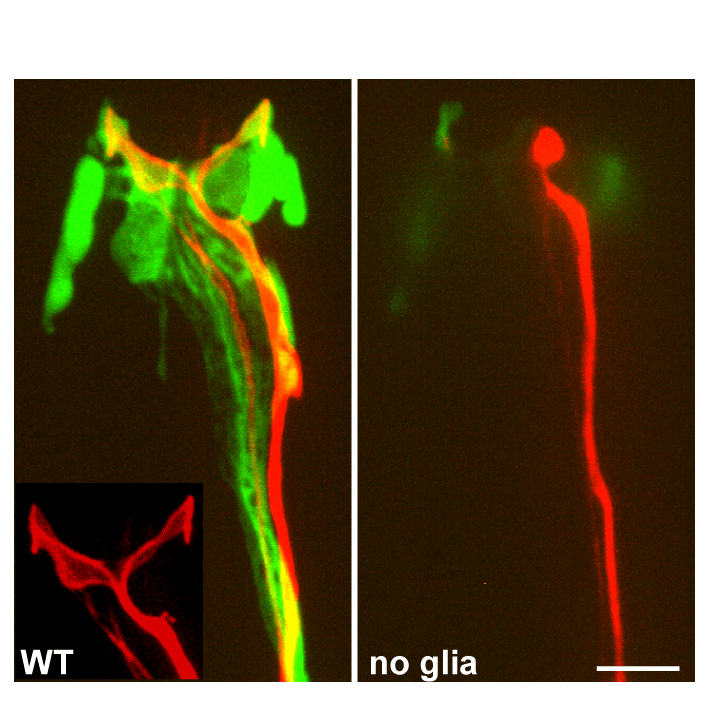 remained full-length; however, neurons failed to function, as manifested by deficits of operated animals in nearly every sensory modality we have tested. By examining the amphid neurons known to be associated with each sensory function we showed that in some cases, defects in neuronal function were accompanied by structural defects at the specialized sensory endings of sensory dendrites which are ensheathed by glia (see Figure). However, in other cases no obvious structural defects were evident by light or electron microscopy, suggesting that glia-secreted factors may regulate neuronal activity. To begin to characterize this putative secreted activity we searched among the glia-enriched genes identified in our microarray studies for glial genes containing signal sequences for secretion. We studied one such gene, fig-1, encoding a protein with several conserved extracellular adhesion domains also present in the protein thrombospondin, a known glial regulator of synaptogenesis. We find that mutations in this gene have no effect on neuronal or glial structure by light or electron microscopy. Remarkably, however, mutants exhibit functional defects in sensory neurons.
These results conclusively demonstrate that amphid glia play an important active role in the regulation of neuronal function.
remained full-length; however, neurons failed to function, as manifested by deficits of operated animals in nearly every sensory modality we have tested. By examining the amphid neurons known to be associated with each sensory function we showed that in some cases, defects in neuronal function were accompanied by structural defects at the specialized sensory endings of sensory dendrites which are ensheathed by glia (see Figure). However, in other cases no obvious structural defects were evident by light or electron microscopy, suggesting that glia-secreted factors may regulate neuronal activity. To begin to characterize this putative secreted activity we searched among the glia-enriched genes identified in our microarray studies for glial genes containing signal sequences for secretion. We studied one such gene, fig-1, encoding a protein with several conserved extracellular adhesion domains also present in the protein thrombospondin, a known glial regulator of synaptogenesis. We find that mutations in this gene have no effect on neuronal or glial structure by light or electron microscopy. Remarkably, however, mutants exhibit functional defects in sensory neurons.
These results conclusively demonstrate that amphid glia play an important active role in the regulation of neuronal function.
Glia as sensory cells: Although most of our studies are geared towards understanding the roles of glia through their interactions with neurons, we have discovered that glia may have neuron-independent sensory roles in C. 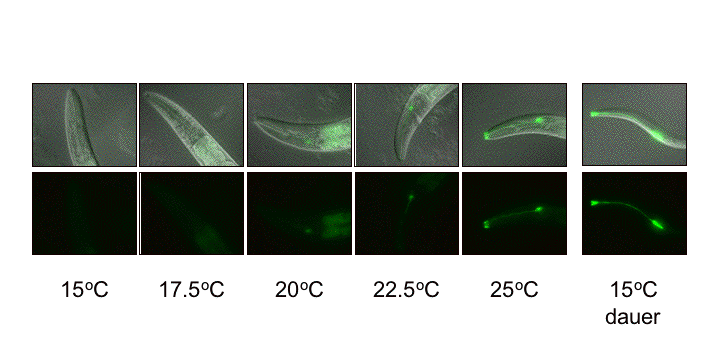 elegans (C. Procko and S. Shaham, manuscript in preparation). We demonstrated that a gene for a cell surface receptor protein is expressed in amphid sheath glia in a temperature-dependent manner, being off at 15C and on at 25C (see Figure). This temperature sensing ability is independent of known thermosensory neurons in C. elegans, as assessed by genetic and cell ablation studies. Temperature-dependent expression of the gene does depend on the TTX-1 transcription factor, known to regulate the function of the AFD amphid thermosensory neuron.
While we are still exploring the meaning and mechanism of this glial temperature-sensing function, our results suggest that glia may have neuron-independent functions in C. elegans. Such functions have not been studied in depth in other animals.
elegans (C. Procko and S. Shaham, manuscript in preparation). We demonstrated that a gene for a cell surface receptor protein is expressed in amphid sheath glia in a temperature-dependent manner, being off at 15C and on at 25C (see Figure). This temperature sensing ability is independent of known thermosensory neurons in C. elegans, as assessed by genetic and cell ablation studies. Temperature-dependent expression of the gene does depend on the TTX-1 transcription factor, known to regulate the function of the AFD amphid thermosensory neuron.
While we are still exploring the meaning and mechanism of this glial temperature-sensing function, our results suggest that glia may have neuron-independent functions in C. elegans. Such functions have not been studied in depth in other animals.
Developmental control of programmed cell death Although much is known regarding the molecular machinery promoting execution of apoptosis, little attention has been given to mechanisms regulating this machinery in individual cells during animal development. We are addressing three interrelated questions: First, we aim to understand the molecular features that distinguish cells destined to live from those destined to die. Second, we aim to understand how temporal control of cell death is achieved. Third, we aim to determine whether caspase-dependent cell death is the only mode of cell death during animal development, or whether alternative death pathways exist. The decision to die: One hypothesis for why certain cells die and others survive during animal development is that cells fated to die express death-promoting proteins not expressed in cells destined to live. We surmised that one such protein class may be the pro-apoptotic BH3 domain-only proteins, since many such proteins exist in vertebrates, 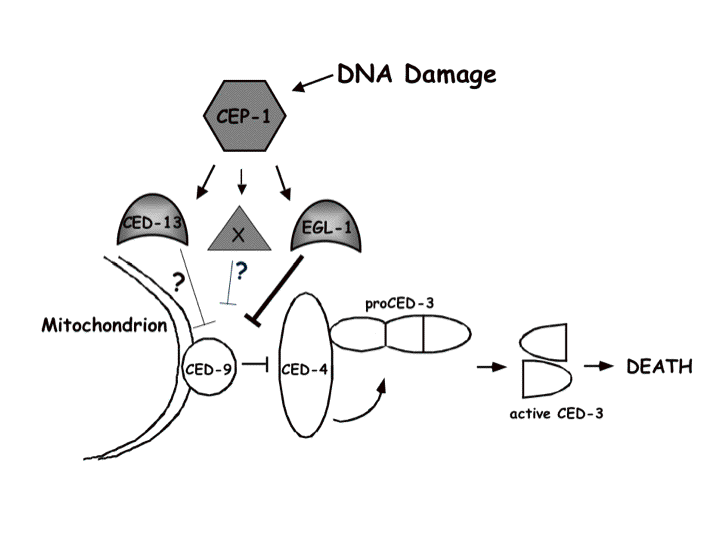 suggesting that they may play cell-specific roles in programmed cell death. In C. elegans, the EGL-1 BH3-domain protein regulates both somatic and DNA damage-induced germ cell death, and is expressed in both cells that live and die, suggesting that it is not a cell-specific determinant of death. We therefore examined the C. elegans sequence database to search for other BH3 domain proteins and identified a new gene we named ced-13 (Schumacher et al., 2005). Overexpression of CED-13 promotes caspase-dependent cell death. CED-13 protein interacts, through its BH3 domain, with the Bcl-2 related protein CED-9, and this interaction is required for cell death to occur. Mutations in ced-13 do not affect somatic or germline developmental cell death, but affect DNA damage-induced cell death. ced-13 mRNA is only detected in the C. elegans germline, and only in response to DNA damage. This regulated transcription absolutely requires the C. elegans p53 gene. Thus, we have identified the first C. elegans cell death gene whose expression is restricted to a subset of dying cells. Furthermore, these studies have contributed to elucidation of the pathway controlling DNA-damage-induced cell death (see Figure).
Remarkably, in C. elegans, two BH3 domain genes, egl-1 and ced-13, are transcriptionally regulated by p53, with egl-1 having a broader role in cell death. Similarly, in vertebrates, two BH3 domain genes, Noxa and PUMA are transcriptionally regulated by p53, with one, PUMA, having a broader cell death role. This apparent conservation suggests that ced-13 and egl-1 may have different functions in response to DNA damage, or that they are regulated by different stimuli.
suggesting that they may play cell-specific roles in programmed cell death. In C. elegans, the EGL-1 BH3-domain protein regulates both somatic and DNA damage-induced germ cell death, and is expressed in both cells that live and die, suggesting that it is not a cell-specific determinant of death. We therefore examined the C. elegans sequence database to search for other BH3 domain proteins and identified a new gene we named ced-13 (Schumacher et al., 2005). Overexpression of CED-13 promotes caspase-dependent cell death. CED-13 protein interacts, through its BH3 domain, with the Bcl-2 related protein CED-9, and this interaction is required for cell death to occur. Mutations in ced-13 do not affect somatic or germline developmental cell death, but affect DNA damage-induced cell death. ced-13 mRNA is only detected in the C. elegans germline, and only in response to DNA damage. This regulated transcription absolutely requires the C. elegans p53 gene. Thus, we have identified the first C. elegans cell death gene whose expression is restricted to a subset of dying cells. Furthermore, these studies have contributed to elucidation of the pathway controlling DNA-damage-induced cell death (see Figure).
Remarkably, in C. elegans, two BH3 domain genes, egl-1 and ced-13, are transcriptionally regulated by p53, with egl-1 having a broader role in cell death. Similarly, in vertebrates, two BH3 domain genes, Noxa and PUMA are transcriptionally regulated by p53, with one, PUMA, having a broader cell death role. This apparent conservation suggests that ced-13 and egl-1 may have different functions in response to DNA damage, or that they are regulated by different stimuli.
Temporal control of cell death onset: Temporal control of programmed cell death is necessary to ensure that cells die only at the right time during animal development. In some C. elegans somatic cells, transcription of egl-1 promotes cell death. EGL-1 protein inhibits the CED-9/Bcl-2 protein, resulting in release of the 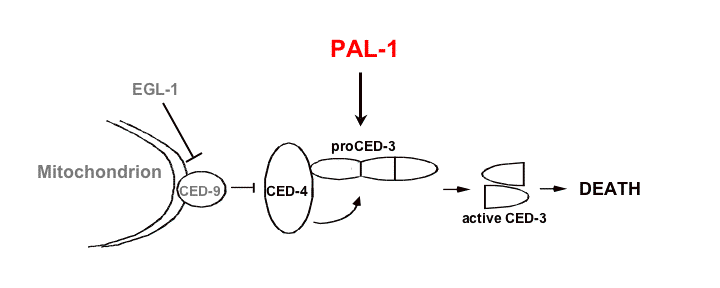 caspase activator CED-4/Apaf-1. Subsequent activation of the CED-3 caspase by CED-4 leads to cell death. Despite the important role of egl-1 transcription in promoting CED-3 activity in cells destined to die, it remains unclear whether temporal control of cell death is mediated by egl-1 expression. Most C. elegans cells that die during somatic development do so within 30-60 minutes after being born, making it difficult to dissect the contributions of individual cell death genes to the control of timing of cell death onset. We therefore chose to study the C. elegans tail-spike cell (Maurer et al., 2007). This binucleate cell dies five hours after it is born, surviving ten times longer than most cells destined to die in C. elegans, thus facilitating analysis of the kinetics of cell death onset. Whereas most dying cells in C. elegans are undifferentiated, the tail-spike cell exhibits extensive differentiated features before dying, including a posterior filamentous process that may function as a scaffold for modeling the C. elegans tail. We have shown that egl-1 and ced-9 play only a minor role in the death of the C. elegans tail-spike cell, demonstrating that temporal control of tail-spike cell death can be achieved in the absence of egl-1. Using GFP reporters and RT-PCR studies we showed that the timing of tail-spike cell death onset is controlled by transcriptional induction of the ced-3 caspase gene. In the tail-spike cell, ced-3 expression is induced minutes before the cell dies, and this induction is sufficient to promote the cellŐs demise. From a genetic screen we isolated three mutants that prevent both ced-3 expression in and death of the tail-spike cell. We showed, using standard cloning strategies, that two of these mutants disrupted a gene encoding the transcription factor PAL-1, the C. elegans homolog of the mammalian intestinal tumor suppressor gene Cdx2. PAL-1 is expressed in the tail-spike cell and can bind in vitro to ced-3 promoter sites critical for tail-spike cell death, suggesting that it promotes cell death by directly activating ced-3 transcription (see Figure).
These results highlight a previously undescribed role for transcriptional regulation of caspases in controlling the timing of cell death onset during animal development, and suggest that cell-specific transcription of caspases may be an important mode of regulating cell-specific death. Furthermore, our results suggest that tumor formation in the absence of Cdx2 function in mammals may be caused, at least in part, by defects in caspase expression. Indeed, in the digestive tract, Cdx2 expression is present at highest level in differentiated epithelial cells that ultimately undergo apoptosis.
caspase activator CED-4/Apaf-1. Subsequent activation of the CED-3 caspase by CED-4 leads to cell death. Despite the important role of egl-1 transcription in promoting CED-3 activity in cells destined to die, it remains unclear whether temporal control of cell death is mediated by egl-1 expression. Most C. elegans cells that die during somatic development do so within 30-60 minutes after being born, making it difficult to dissect the contributions of individual cell death genes to the control of timing of cell death onset. We therefore chose to study the C. elegans tail-spike cell (Maurer et al., 2007). This binucleate cell dies five hours after it is born, surviving ten times longer than most cells destined to die in C. elegans, thus facilitating analysis of the kinetics of cell death onset. Whereas most dying cells in C. elegans are undifferentiated, the tail-spike cell exhibits extensive differentiated features before dying, including a posterior filamentous process that may function as a scaffold for modeling the C. elegans tail. We have shown that egl-1 and ced-9 play only a minor role in the death of the C. elegans tail-spike cell, demonstrating that temporal control of tail-spike cell death can be achieved in the absence of egl-1. Using GFP reporters and RT-PCR studies we showed that the timing of tail-spike cell death onset is controlled by transcriptional induction of the ced-3 caspase gene. In the tail-spike cell, ced-3 expression is induced minutes before the cell dies, and this induction is sufficient to promote the cellŐs demise. From a genetic screen we isolated three mutants that prevent both ced-3 expression in and death of the tail-spike cell. We showed, using standard cloning strategies, that two of these mutants disrupted a gene encoding the transcription factor PAL-1, the C. elegans homolog of the mammalian intestinal tumor suppressor gene Cdx2. PAL-1 is expressed in the tail-spike cell and can bind in vitro to ced-3 promoter sites critical for tail-spike cell death, suggesting that it promotes cell death by directly activating ced-3 transcription (see Figure).
These results highlight a previously undescribed role for transcriptional regulation of caspases in controlling the timing of cell death onset during animal development, and suggest that cell-specific transcription of caspases may be an important mode of regulating cell-specific death. Furthermore, our results suggest that tumor formation in the absence of Cdx2 function in mammals may be caused, at least in part, by defects in caspase expression. Indeed, in the digestive tract, Cdx2 expression is present at highest level in differentiated epithelial cells that ultimately undergo apoptosis.
Discovery of a novel non-apoptotic morphologically conserved developmental cell death pathway: To further understand the temporal control of cell death and to identify additional mechanisms regulating cell-specific cell death in C. elegans we have also studied the death of the male-specific linker cell (Abraham et al., 2005). 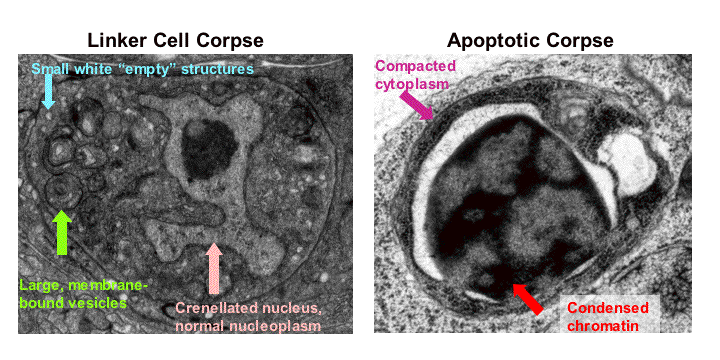 The death of the linker cell, two days after it is born, during or just after the L4/adult transition, may facilitate fusion between the gonad and cloaca, connecting the male reproductive system to the exterior. Ablation of cells neighboring the linker cell, and examination of mutants in which the linker cell migrates abnormally support the notion that the linker cell employs a cell-autonomous program to promote its demise. These studies we performed also demonstrate that a local cue regulating the efficiency of linker cell death is present in the cloacal region. We found that linker cell death is controlled by the microRNA let-7 and by the Zn-finger transcription factor LIN-29, both components of the main developmental timing pathway in C. elegans, suggesting that the temporal cue promoting linker cell death may be mediated by this pathway. LIN-29 functions within the linker cell to promote its demise, consistent with the idea that the linker cell employs a cell-autonomous death program. A map kinase cascade function in parallel to LIN-29 to promote linker cell demise (Elyse Blum, unpublished results). To determine which cell death genes may be targets of LIN-29 we examined animals carrying mutations in these genes. Remarkably, we discovered that linker cell death is completely independent of all known C. elegans cell death genes, including the C. elegans caspase ced-3 and all other caspase-related genes. Linker cell death proceeds efficiently even in animals carrying multiple mutations that block the death of other C. elegans cells destined to die. Furthermore, although the linker cell can be engulfed by the canonical apoptotic cell engulfment program when the cell is at an abnormal position in the animal, a previously undescribed engulfment program is required for its clearance at its normal location. Electron microscopy of dying linker cells reveals non-apoptotic features including nuclear crenellation in the absence of chromatin condensation, swelling of mitochondria and endoplasmic reticulum, and accumulation of single-and multi-layered cytoplasmic membrane-bound structures (see Figure). Strikingly similar morphological changes occur during the normal developmental death of neurons in the vertebrate spinal cord and ciliary ganglia, suggesting that the molecular basis of this non-canonical cell death program may be conserved.
Although non-apoptotic, caspase-independent programmed cell death pathways have been postulated, little is known about their molecular constituents or possible in vivo functions. We have, thus, for the first time, provided proof of the in vivo existence of such a pathway, and are well-poised to uncover its genetic basis. Despite the pervasiveness of cell death during mammalian development, mutations in known apoptosis genes in the mouse fail to block the gross progress of development. Furthermore, in the immune system of mouse apoptosis mutants, the total number of cells is not increased to an extent that might be predicted given the large numbers of cells eliminated during normal T and B cell development. Although these observations can be explained by feedback control of cell numbers, and/or redundancy within caspase and other cell death gene families, it is also possible that these cell deaths are regulated by an entirely different pathway similar to the one regulating linker cell death. If this is the case, this alternative mode of cell death must, therefore, play a major role during normal vertebrate development.
The death of the linker cell, two days after it is born, during or just after the L4/adult transition, may facilitate fusion between the gonad and cloaca, connecting the male reproductive system to the exterior. Ablation of cells neighboring the linker cell, and examination of mutants in which the linker cell migrates abnormally support the notion that the linker cell employs a cell-autonomous program to promote its demise. These studies we performed also demonstrate that a local cue regulating the efficiency of linker cell death is present in the cloacal region. We found that linker cell death is controlled by the microRNA let-7 and by the Zn-finger transcription factor LIN-29, both components of the main developmental timing pathway in C. elegans, suggesting that the temporal cue promoting linker cell death may be mediated by this pathway. LIN-29 functions within the linker cell to promote its demise, consistent with the idea that the linker cell employs a cell-autonomous death program. A map kinase cascade function in parallel to LIN-29 to promote linker cell demise (Elyse Blum, unpublished results). To determine which cell death genes may be targets of LIN-29 we examined animals carrying mutations in these genes. Remarkably, we discovered that linker cell death is completely independent of all known C. elegans cell death genes, including the C. elegans caspase ced-3 and all other caspase-related genes. Linker cell death proceeds efficiently even in animals carrying multiple mutations that block the death of other C. elegans cells destined to die. Furthermore, although the linker cell can be engulfed by the canonical apoptotic cell engulfment program when the cell is at an abnormal position in the animal, a previously undescribed engulfment program is required for its clearance at its normal location. Electron microscopy of dying linker cells reveals non-apoptotic features including nuclear crenellation in the absence of chromatin condensation, swelling of mitochondria and endoplasmic reticulum, and accumulation of single-and multi-layered cytoplasmic membrane-bound structures (see Figure). Strikingly similar morphological changes occur during the normal developmental death of neurons in the vertebrate spinal cord and ciliary ganglia, suggesting that the molecular basis of this non-canonical cell death program may be conserved.
Although non-apoptotic, caspase-independent programmed cell death pathways have been postulated, little is known about their molecular constituents or possible in vivo functions. We have, thus, for the first time, provided proof of the in vivo existence of such a pathway, and are well-poised to uncover its genetic basis. Despite the pervasiveness of cell death during mammalian development, mutations in known apoptosis genes in the mouse fail to block the gross progress of development. Furthermore, in the immune system of mouse apoptosis mutants, the total number of cells is not increased to an extent that might be predicted given the large numbers of cells eliminated during normal T and B cell development. Although these observations can be explained by feedback control of cell numbers, and/or redundancy within caspase and other cell death gene families, it is also possible that these cell deaths are regulated by an entirely different pathway similar to the one regulating linker cell death. If this is the case, this alternative mode of cell death must, therefore, play a major role during normal vertebrate development.
Technology development Temporal Control of Cell-Specific Transgene Expression: Cell-specific promoters allow only spatial control of transgene expression in C. elegans. We developed a method, using cell-specific rescue of heat-shock factor-1 (hsf-1) mutants, that allows spatial and temporal regulation of 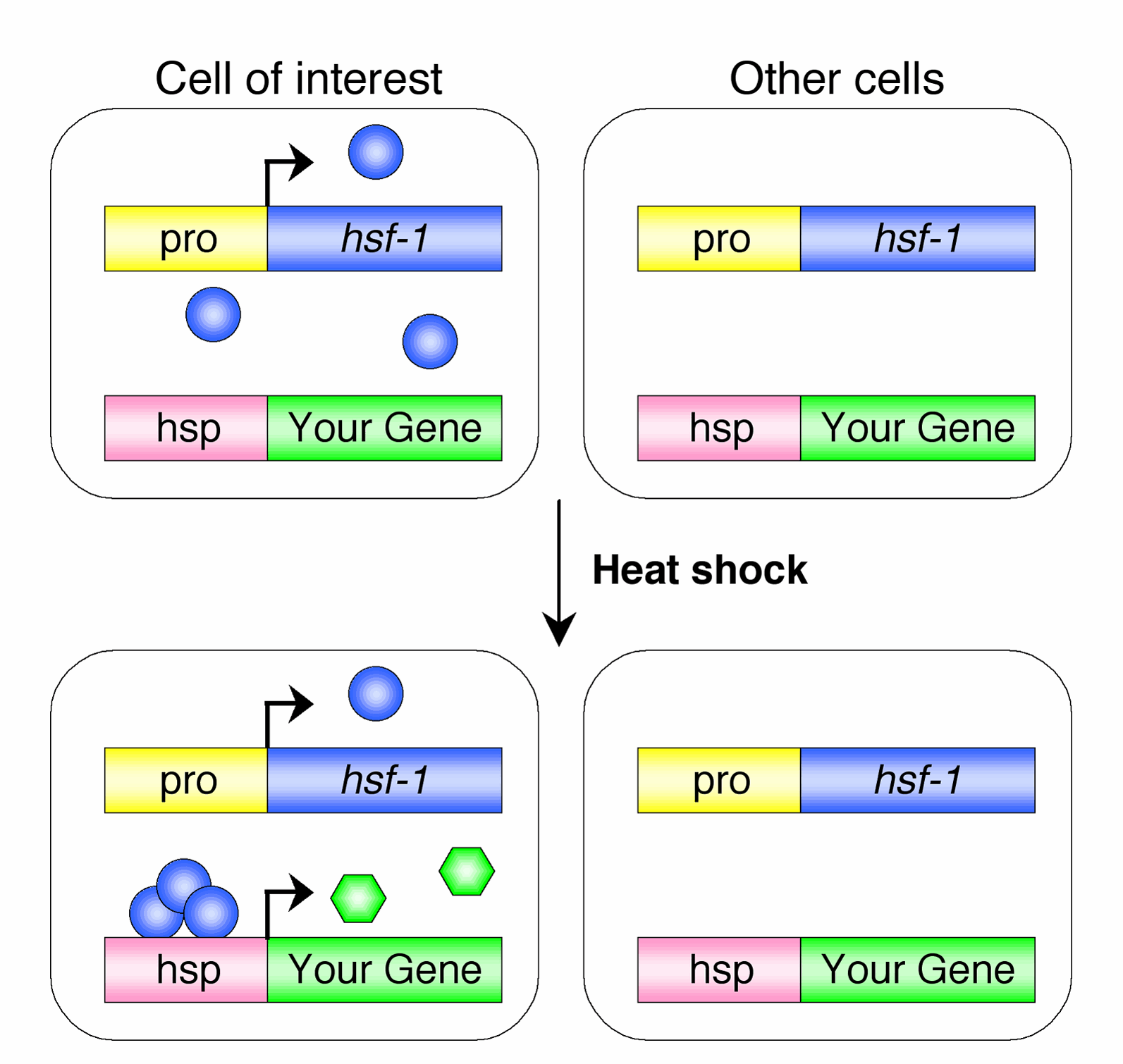 transgene expression (Bacaj and Shaham, 2007). We demonstrated the utility of this method for timed
reporter gene expression and for temporal studies of gene function (see Figure). We transformed hsf-1(sy441)
mutants with two transgenes simultaneously. The first transgene, referred to as the driver, consisted of the hsf-1 cDNA under the control of the 5-kb promoter region of the gene vap-1, which is expressed specifically within the amphid sheath glial cell. The second transgene, termed the responder, consisted of the gene encoding GFP under the control of the 400-bp promoter region of the hsp-16.2 gene. hsf-1(sy441) animals carrying both transgenes as an extrachromosomal array, and raised at 20C, did not display detectable GFP expression. However, following administration of a heat-shock at 34C for 30 min, GFP expression was observed specifically within the amphid sheath cells. GFP fluorescence was visible within 1 h following the temperature shift and was still evident after 24 h. We have now demonstrated the utility of this method to at least three different promoter/cell combinations.
transgene expression (Bacaj and Shaham, 2007). We demonstrated the utility of this method for timed
reporter gene expression and for temporal studies of gene function (see Figure). We transformed hsf-1(sy441)
mutants with two transgenes simultaneously. The first transgene, referred to as the driver, consisted of the hsf-1 cDNA under the control of the 5-kb promoter region of the gene vap-1, which is expressed specifically within the amphid sheath glial cell. The second transgene, termed the responder, consisted of the gene encoding GFP under the control of the 400-bp promoter region of the hsp-16.2 gene. hsf-1(sy441) animals carrying both transgenes as an extrachromosomal array, and raised at 20C, did not display detectable GFP expression. However, following administration of a heat-shock at 34C for 30 min, GFP expression was observed specifically within the amphid sheath cells. GFP fluorescence was visible within 1 h following the temperature shift and was still evident after 24 h. We have now demonstrated the utility of this method to at least three different promoter/cell combinations.
Optimizing genetic screens in C. elegans: In genetic screens, the number of mutagenized gametes examined is an important parameter for evaluating screen progress, the number of genes of a given mutable phenotype, gene 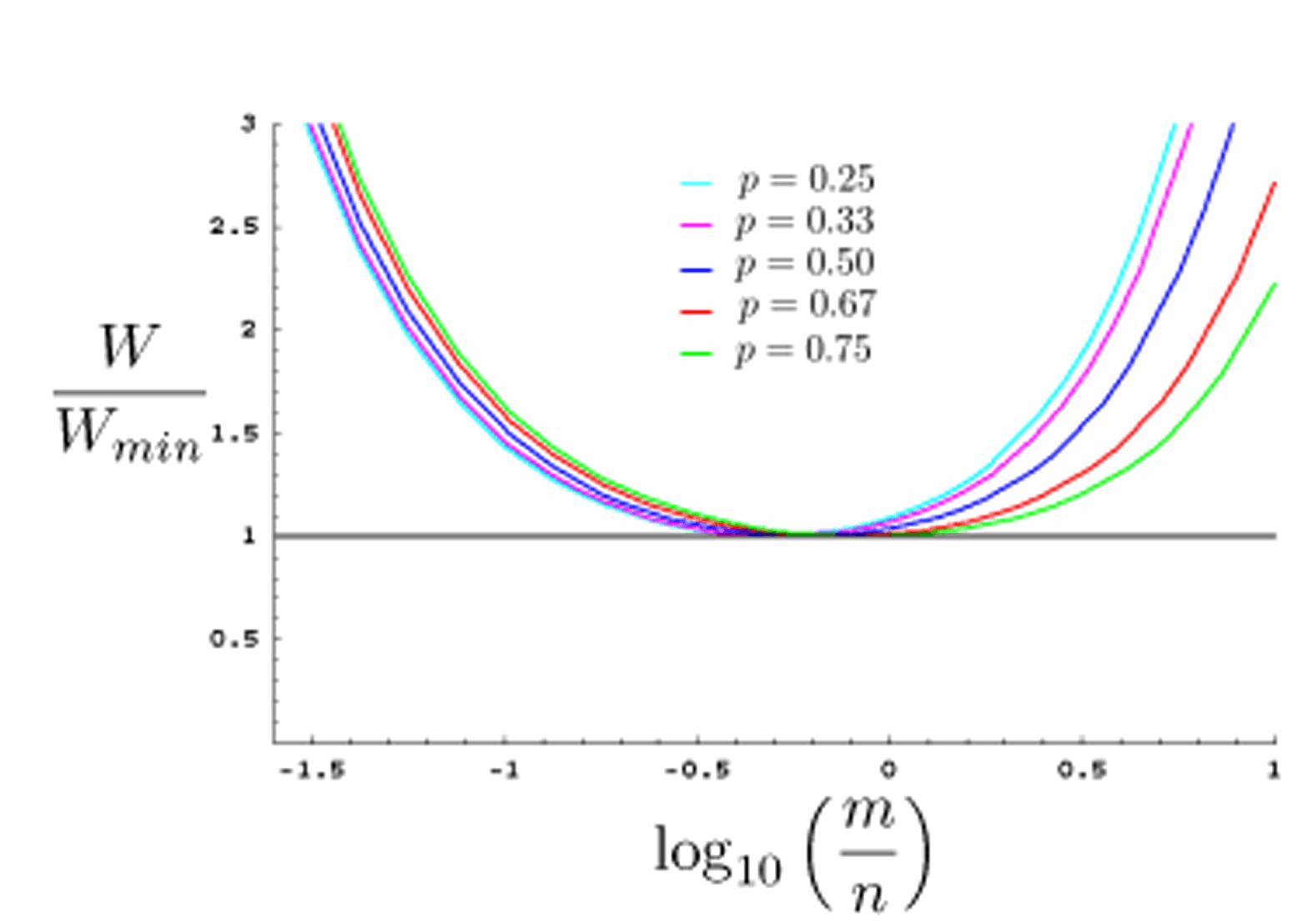 size, cost, and labor. Since genetic screens often entail examination
of thousands or tens of thousands of animals, strategies for optimizing genetics screens are important for minimizing effort
while maximizing the number of mutagenized gametes examined. Such strategies have not been described for
genetic screens in C. elegans. We have described general principles of genetic screens in C. elegans,
and use a modified binomial strategy to obtain a general expression for the number of mutagenized gametes examined in
a genetic screen. We used this expression to calculate optimal screening parameters for a large range of genetic screen types (Shaham, 2007). In
addition, we developed a simple online genetic-screen-optimization tool that can be used independently of this paper. Our
results demonstrate that choosing the optimal F2-to-F1 screening ratio (see Figure) can significantly improve screen efficiency.
size, cost, and labor. Since genetic screens often entail examination
of thousands or tens of thousands of animals, strategies for optimizing genetics screens are important for minimizing effort
while maximizing the number of mutagenized gametes examined. Such strategies have not been described for
genetic screens in C. elegans. We have described general principles of genetic screens in C. elegans,
and use a modified binomial strategy to obtain a general expression for the number of mutagenized gametes examined in
a genetic screen. We used this expression to calculate optimal screening parameters for a large range of genetic screen types (Shaham, 2007). In
addition, we developed a simple online genetic-screen-optimization tool that can be used independently of this paper. Our
results demonstrate that choosing the optimal F2-to-F1 screening ratio (see Figure) can significantly improve screen efficiency.
galign: a Tool for Rapid Genome Polymorphism Discovery: Highly parallel sequencing technologies have become important tools in the analysis of sequence polymorphisms on a genomic scale. However, the development of customized software to analyze data produced by these methods has lagged behind. We developed a tool, 'galign', 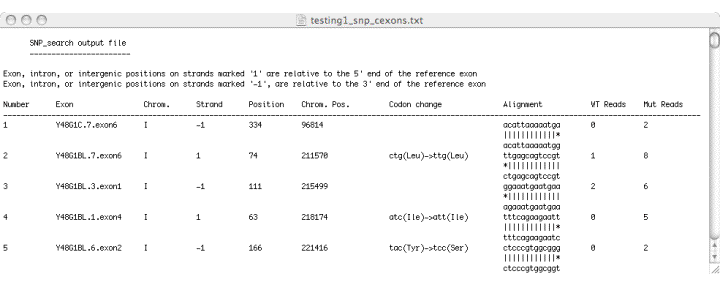 designed to identify polymorphisms between sequence
reads obtained using Illumina/Solexa technology and a reference genome. The 'galign'
alignment tool does not use Smith-Waterman matrices for sequence comparisons.
Instead, a simple algorithm comparing parsed sequence reads to parsed reference genome
sequences is used. 'galign' output is geared towards immediate user application,
displaying polymorphism locations, nucleotide changes, and relevant predicted aminoacid
changes for ease of information processing. To do so, 'galign' requires several
accessory files easily derived from an annotated reference genome. Direct sequencing as
well as in silico studies demonstrate that 'galign' provides lesion predictions comparable
in accuracy to available prediction programs, and accompanied by greater processing
speed and more user-friendly output. We demonstrate the use of 'galign' to identify
mutations leading to phenotypic consequences in C. elegans.
Our studies suggest that 'galign' is a useful tool for polymorphism discovery, and is of
immediate utility for sequence mining in C. elegans.
designed to identify polymorphisms between sequence
reads obtained using Illumina/Solexa technology and a reference genome. The 'galign'
alignment tool does not use Smith-Waterman matrices for sequence comparisons.
Instead, a simple algorithm comparing parsed sequence reads to parsed reference genome
sequences is used. 'galign' output is geared towards immediate user application,
displaying polymorphism locations, nucleotide changes, and relevant predicted aminoacid
changes for ease of information processing. To do so, 'galign' requires several
accessory files easily derived from an annotated reference genome. Direct sequencing as
well as in silico studies demonstrate that 'galign' provides lesion predictions comparable
in accuracy to available prediction programs, and accompanied by greater processing
speed and more user-friendly output. We demonstrate the use of 'galign' to identify
mutations leading to phenotypic consequences in C. elegans.
Our studies suggest that 'galign' is a useful tool for polymorphism discovery, and is of
immediate utility for sequence mining in C. elegans.
|